E5 Timepoint 1, 2, 3, and 4 Instantaneous Calcification Sample Processing (Samples Collected in 2020)
Processing the E5 January, March, September, and November 2020 Instantaneous Calcification Samples
Goal
Process calcification samples collected for the E5 uROL Epigenetics Project from January, March, September, and November 2020 (timepoints 1, 2, 3, and 4). The samples were collected in January (n = 146), March (n = 151), September 2020 (n = 114), and November 2020 (n = 145) and fixed with 75uL of saturated mercuric chloride for transportation back to URI on November, 22nd 2020. All samples were brought back from the January and March timepoints November 2020, but only 51 bottles could fit in the containers from the September timepoint. The remaining samples for the September and November 2020 timepoints are as follows: the Silbiger lab brought back the remaining samples from Mo’orea for the September 2020 timepoint in August 2021 and processed them on their titrator in October 2021 following the Silbiger Lab Titrator Protocols. The November 2020 samples were brought back to URI by Hollie and Ariana in November 2021. All remaining samples are in the URI CBLS 180 Environmental Room.
Samples Remaining
- January 2020, 146 samples remaining
- September 2020, 51 samples remaining
- November 2020, 145 samples remaining
Process
Samples were collected, experimental run, and transported back to URI following the E5 Instantaneous Calcification Protocol in Mo’orea, French Polynesia.
Upon the return to URI, we wanted to test the total alkalinity signal we would receive from our initial seawater samples before the experimental procedure and after to calculate delta total alkalinity. To do so, titrations will need to be completed for all samples to calculate total alkalinity following the alkalinity anomaly technique (Chisholm and Gattuso, 1991).
All samples will be processed following the established Putnam Lab Titrator Protocol.
All data processed and collected on the Putnam Lab Titrator will be available on the Putnam Lab Titrator GitHub Repository.
Titration Data Entry Process
- After uploading titration data to the Putnam Lab Titrator Repository from the CBLS lab computer, you will need to enter this into the E5 uROL Epigenetics Project Timeseries Data.
- First, copy the titrator TA outputs for each sample into the respective TP1, TP2, TP3, and TP4 data sheets found in the E5 uROL Epigenetics Project Timeseries Data Page.
You can find the information highlighted in green from the labels on the bottles, and the rest of the information can be found using the instructions below
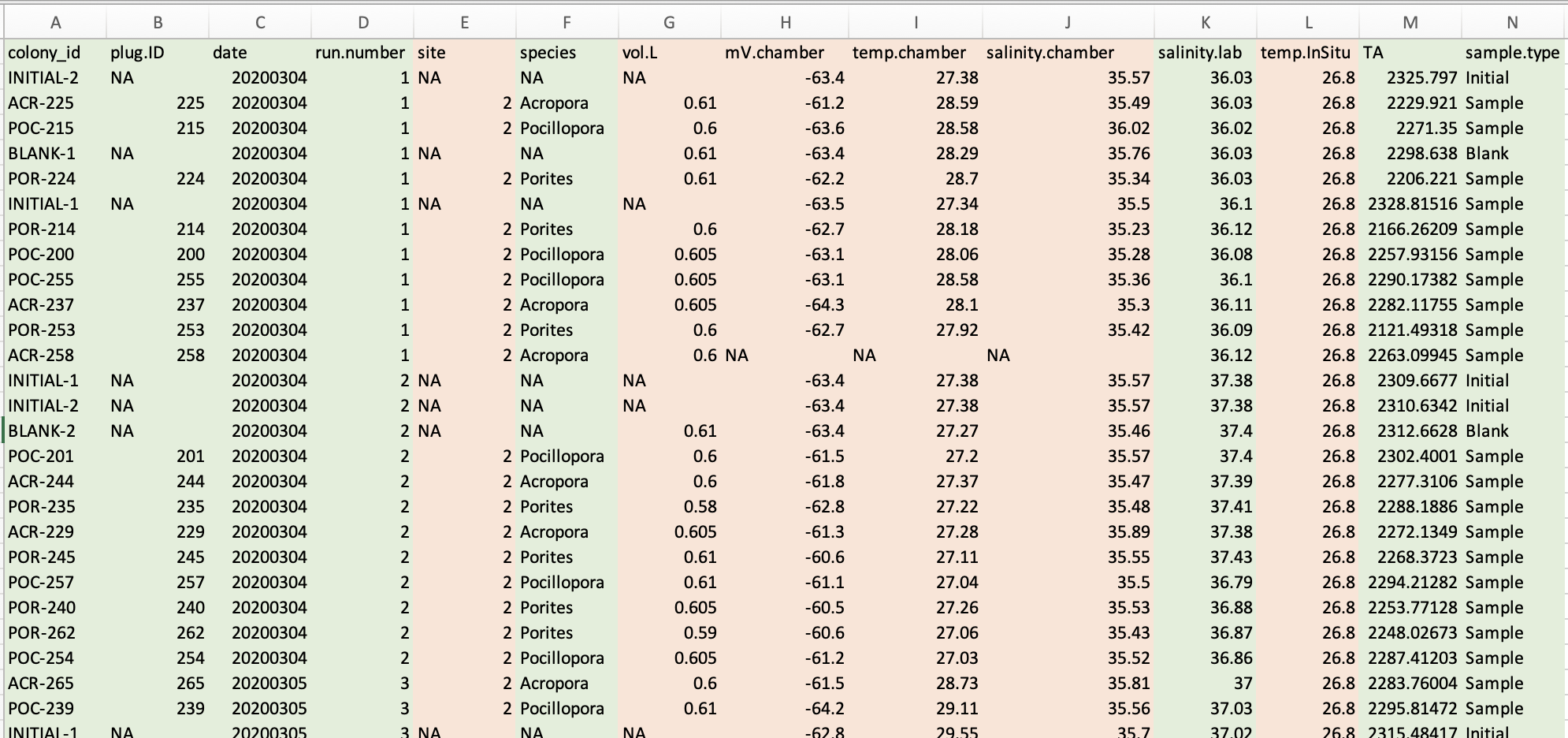
- Second, you will need to match the corresponding Mo’orea measurements (i.e. lab pH, volume, salinity, and temperature in Mo’orea) and information into these data sheets as well.
- The information for this can be found in each individual timeseries 1, 2, 3, and 4 folders on the E5 uROL Epigenetics Project Timeseries Data Page.
- To find this corresponding information for specific samples and blanks, you will look at the 1_DeltaTA_metadata.csv, 2_DeltaTA_metadata.csv, 3_DeltaTA_metadata.csv, and 4_DeltaTA_metadata.csv data sheets.
- To find this information for the initial water samples, you will look at the 2_initial_TA_samples.csv, 3_initial_TA_samples.csv, and 4_initial_TA_samples.csv data sheets. I have not been able to find the initial sample data sheet for TP1 so you may need to ask Ariana and/or Hollie.
- I have completed all of the samples for timepoint 2 (March 2020) so if you want to see data examples for comparison that is a good place to look.
Data and notes for testing pH probe and buffers on 20210221
Notes
- In the initial titration set-up I noticed that the pH calibration was a bit off due to the zero point and slope values we received here. I ran a CRM and found that is was within the < 1% accuracy that we would expect but at a value of -0.82% was a little high especially with the floating pH calibration data. I began to troubleshoot the problem and thought that maybe the pH buffers could be off from prior knowledge. A new round of buffers was ordered to test the pH probe. pH calibration and CRM accuracy information can be found in the Putnam Lab Titrator Protocol.
Data
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 1791.44566335087 | 59.81 | 35 |
| CRM 1 | 2215.55476501538 | 59.561 | 33.417 |
Data and notes for testing variability in CRMs and pH probe 20210228
Notes
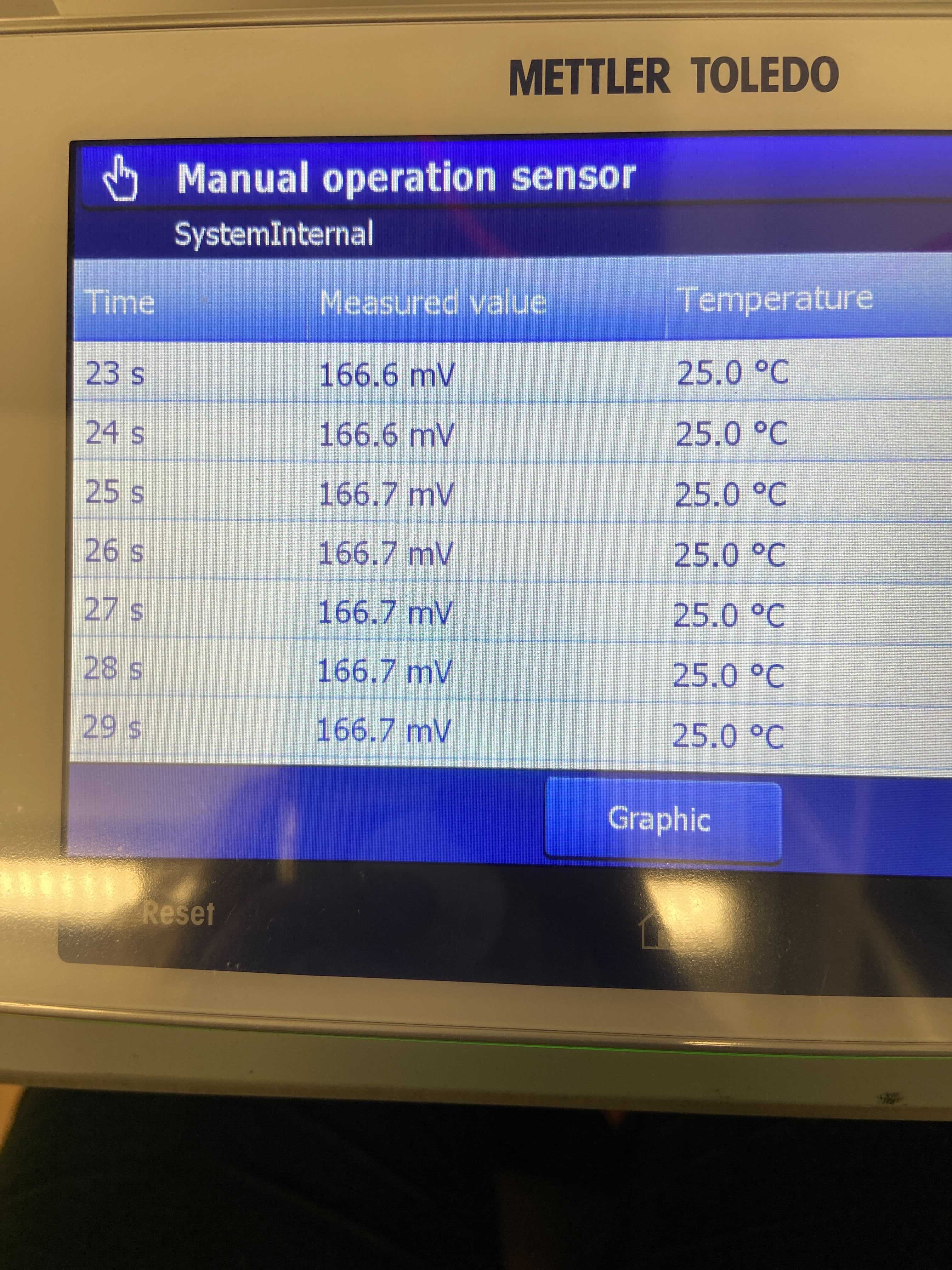
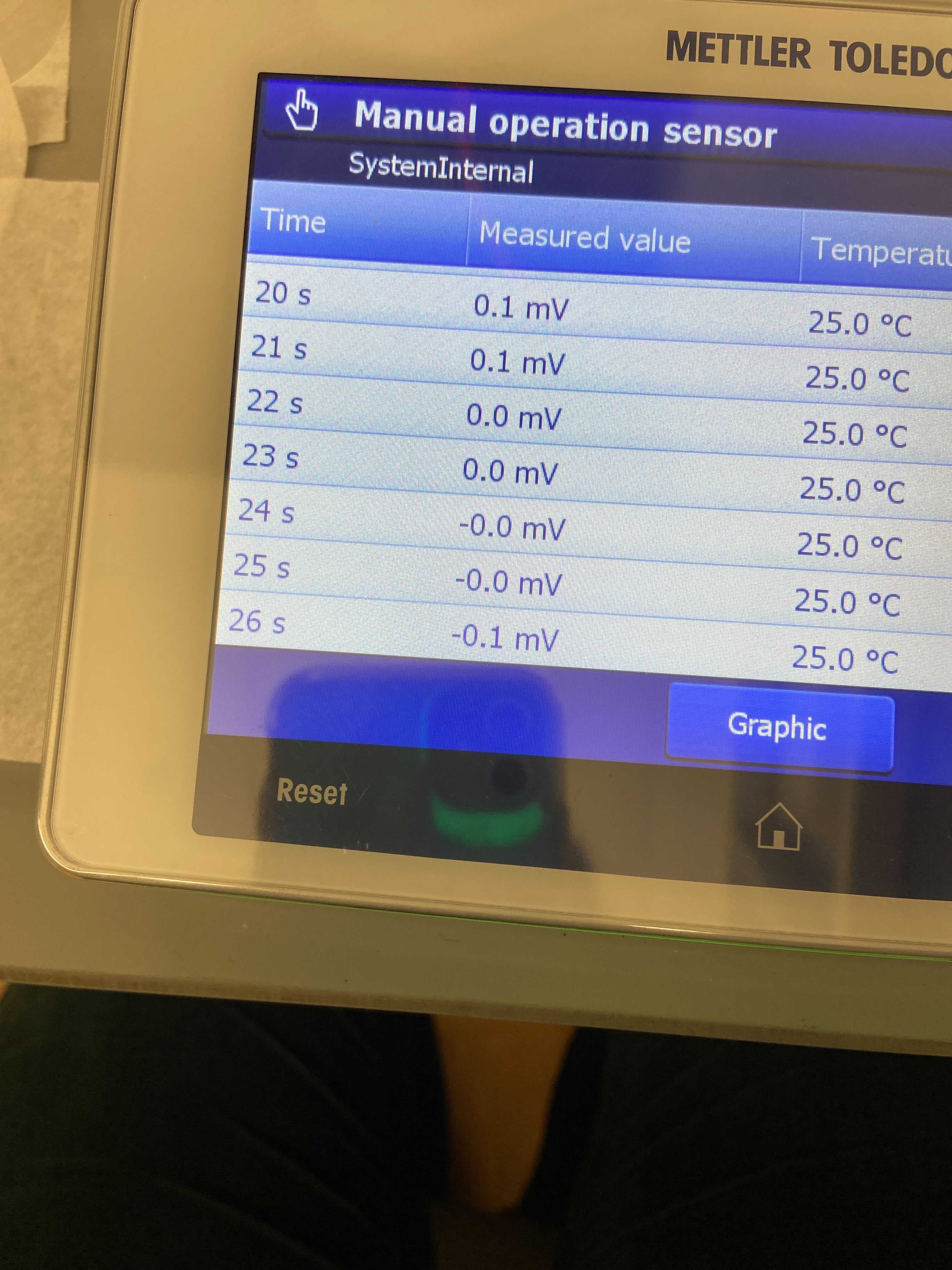
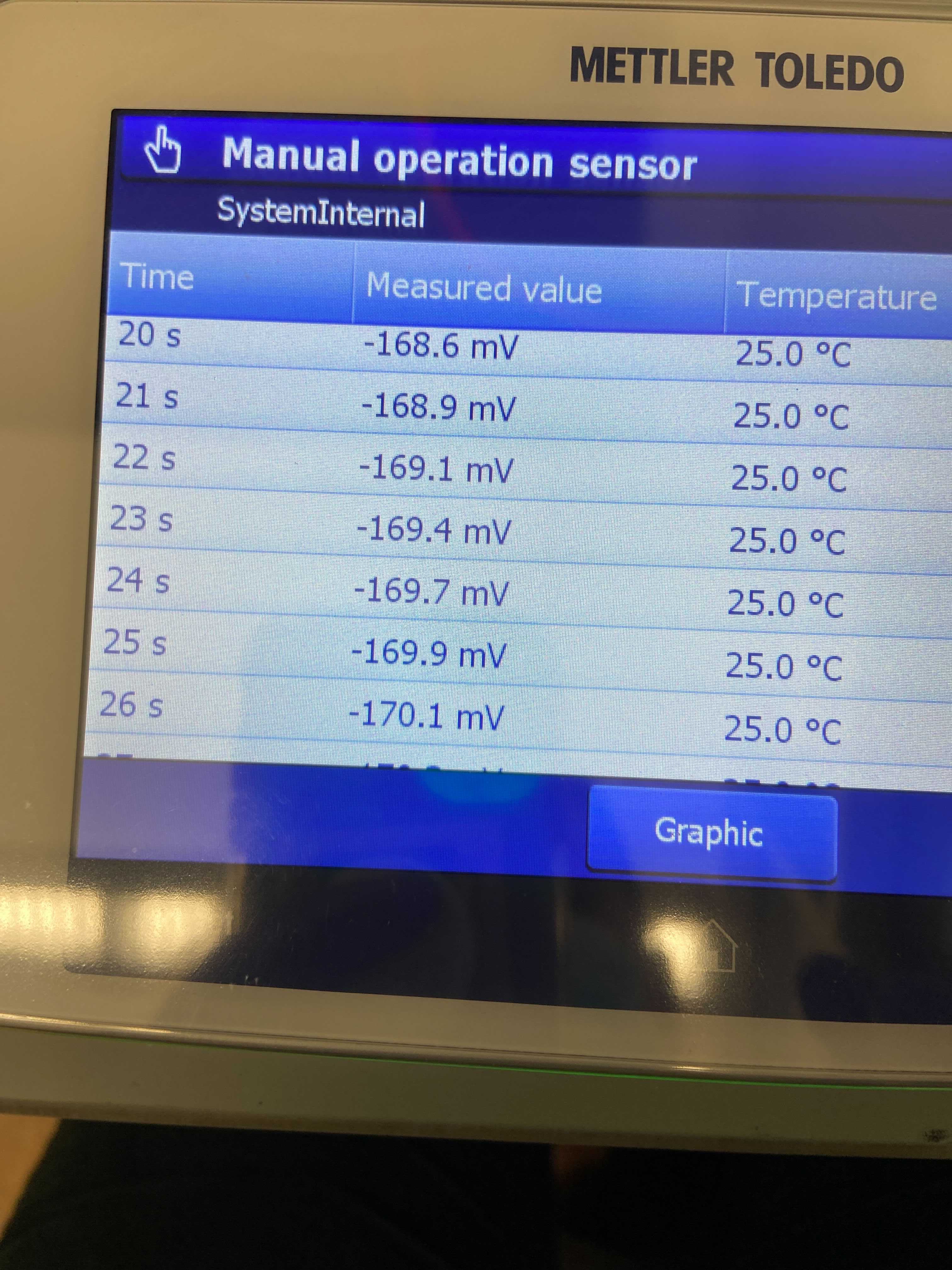
- Continued the troubleshooting process and still saw that the pH calibration values were off and the probe was floating in its mV values when measured manually for 40 seconds in each of the 4, 7, and 10 pH buffers. The values were small but still seemed inconsistent. To test how much variability we were seeing in CRMs due to the pH probe, I ran 8 CRMs. There was a lot of variability between the CRMs so a new pH probe was ordered and received on 20210304.
pH buffer 4 manually measured values (mV)
pH buffer 7 manually measured values (mV)
pH buffer 10 manually measured values (mV)
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210218 | 2215.547681 | 2234.07 | -0.829084093 | 141 | CRM opened 20210216 |
| 20210221 | 2215.554 | 2234.07 | -0.828801246 | 141 | CRM opened 20210221 |
| 20210228 | 2211.139264 | 2234.07 | -1.026410826 | 141 | CRM opened 20210216 |
| 20210228 | 2213.599263 | 2234.07 | -0.916297939 | 141 | CRM opened 20210216 |
| 20210228 | 2218.811032 | 2234.07 | -0.683012077 | 141 | CRM opened 20210228 |
| 20210228 | 2208.830425 | 2234.07 | -1.129757569 | 141 | CRM opened 20210228 |
| 20210228 | 2203.497378 | 2234.07 | -1.368472003 | 141 | CRM opened 20210228 |
| 20210228 | 2213.354976 | 2234.07 | -0.927232556 | 141 | CRM opened 20210228 |
| 20210228 | 2230.94523 | 2234.07 | -0.13986896 | 141 | CRM opened 20210228 |
| 20210228 | 2222.545765 | 2234.07 | -0.515840388 | 141 | CRM opened 20210228 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 932.4357056 | 59.701 | 35 |
| CRM1 | 2211.139264 | 59.861 | 33.417 |
| CRM2 | 2213.599263 | 60.436 | 33.417 |
| CRM3 | 2218.811032 | 60.178 | 33.417 |
| CRM4 | 2208.830425 | 60.307 | 33.417 |
| CRM5 | 2203.497378 | 60.315 | 33.417 |
| CRM6 | 2213.354976 | 60.201 | 33.417 |
| CRM7 | 2230.94523 | 60.413 | 33.417 |
| CRM8 | 2222.545765 | 60.105 | 33.417 |
New pH probe received on 20210303
New pH probe assembled, installed and tested on 20210307
Data and notes for testing variability in CRMs and new pH probe 20210314
Notes
- Continued the troubleshooting process and had great pH calibration values with the new probe and buffers before any runs today. Purged all bubbles and checked every box before running one junk and 8 CRM samples. After the run with 8 CRMS, I noticed that the variability from the CRM value was still >1% in a few of the samples. I attributed this to the older CRM bottle that was opened on 20210228. I opened a brand new CRM bottle and ran four samples but there was still variability to the original CRM value but less than in the previous run. I called Mettler Toledo support and they said this could be an offset with the titrant (acid) and they have had this happen. We could measure the titrant against tris buffer to calculate the offset and apply that to our equations. Noticed that the titrant was replaced in May 2019, which seemed very old and think evaporation could have effected its composition. Next round I am going to change out the titrant with a new bottle.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210314 | 2182.69197 | 2234.07 | -2.29975022 | 141 | CRM opened 20210228 |
| 20210314 | 2183.070705 | 2234.07 | -2.282797543 | 141 | CRM opened 20210228 |
| 20210314 | 2179.989317 | 2234.07 | -2.420724647 | 141 | CRM opened 20210228 |
| 20210314 | 2180.118916 | 2234.07 | -2.414923637 | 141 | CRM opened 20210228 |
| 20210314 | 2194.262889 | 2234.07 | -1.781820203 | 141 | CRM opened 20210228 |
| 20210314 | 2194.808799 | 2234.07 | -1.757384566 | 141 | CRM opened 20210228 |
| 20210314 | 2192.134088 | 2234.07 | -1.877108222 | 141 | CRM opened 20210228 |
| 20210314 | 2192.253611 | 2234.07 | -1.871758246 | 141 | CRM opened 20210228 |
| 20210314 | 2205.731211 | 2224.47 | -0.842393439 | 180 | CRM opened 20210314 |
| 20210314 | 2194.005521 | 2224.47 | -1.369516288 | 180 | CRM opened 20210314 |
| 20210314 | 2192.044753 | 2224.47 | -1.457661693 | 180 | CRM opened 20210314 |
| 20210314 | 2196.378938 | 2224.47 | -1.262820467 | 180 | CRM opened 20210314 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 1981.003448 | 60.113 | 35 |
| CRM1 | 2182.69197 | 60.225 | 33.417 |
| CRM2 | 2183.070705 | 59.774 | 33.417 |
| CRM3 | 2179.989317 | 60.107 | 33.417 |
| CRM4 | 2180.118916 | 60.161 | 33.417 |
| CRM5 | 2194.262889 | 59.659 | 33.417 |
| CRM6 | 2194.808799 | 59.719 | 33.417 |
| CRM7 | 2192.134088 | 59.651 | 33.417 |
| CRM8 | 2192.253611 | 59.892 | 33.417 |
| CRM1 | 2205.731211 | 60.297 | 33.623 |
| CRM2 | 2194.005521 | 60.319 | 33.623 |
| CRM3 | 2192.044753 | 59.881 | 33.623 |
| CRM4 | 2196.378938 | 59.971 | 33.623 |
Data and notes for testing variability in CRMs with new titrant 20210328
Notes
- Continued the troubleshooting process and changed out the older titrant with a new Batch #A16 titrant. Purged the lines four times to make sure to run through any of the older titrant from the lines. Ran a pH calibration that had sound numbers and then I ran a junk sample before five CRMs. IT WORKED! The CRM values were very accurate and everything looked good from the run. Can continue with samples moving forward.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210328 | 2766.991908 | 2225.47 | 24.33292331 | 180 | CRM opened 20210314 |
| 20210328 | 2226.220045 | 2226.47 | -0.011226509 | 180 | CRM opened 20210314 |
| 20210328 | 2224.425627 | 2227.47 | -0.136674041 | 180 | CRM opened 20210314 |
| 20210328 | 2224.201971 | 2228.47 | -0.191522852 | 180 | CRM opened 20210314 |
| 20210328 | 2225.549637 | 2229.47 | -0.175842846 | 180 | CRM opened 20210314 |
| 20210328 | 2228.292488 | 2230.47 | -0.097625722 | 180 | CRM opened 20210314 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 2766.9919084924 | 60.137 | 35 |
| CRM1 | 2226.22004515256 | 60.18 | 33.623 |
| CRM2 | 2224.4256267384 | 60.274 | 33.623 |
| CRM3 | 2224.20197069824 | 60.29 | 33.623 |
| CRM4 | 2225.54963650217 | 60.077 | 33.623 |
| CRM5 | 2228.2924875591 | 60.135 | 33.623 |
Data and notes for testing E5 samples TA signal 20210504
Notes
- The CRM values were very accurate and everything looked good from the previous run, testing a batch of samples from March (TP2) run 1 on 20200304 to see if there is a processable deltaTA signal from the initial sample to the blank and other coral sample chambers. All went well and we were able to see > 30 for deltaTA in almost all samples which is a good value moving forward. Going to run more samples from another run to make sure this occurs in other replicates.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210504 | 2224.823 | 2224.47 | 0.015869 | 180 | CRM opened 20210418 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 2030.80445223374 | 60.035 | 35 |
| INITIAL-2 | 2325.79665116253 | 59.525 | 36.03 |
| ACR-225 | 2229.92073279084 | 60.488 | 36.03 |
| POC-215 | 2271.34957005683 | 59.695 | 36.02 |
| BLANK-1 | 2298.63819200678 | 60.297 | 36.03 |
| POR-224 | 2206.22121318086 | 60.266 | 36 |
Data and notes for testing E5 samples TA signal 20210506
Notes
- The CRM values were very accurate and everything looked good from the previous run, testing a batch of samples from March (TP2) run 2 on 20200304 to see if the deltaTA values are also representative of what we saw in yesterdays runs. Most of the samples again had deltaTA’s > 30 and all looked good.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210506 | 2225.094 | 2224.47 | 0.028052 | 180 | CRM opened 20210418 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 2018.39880477983 | 60.226 | 35 |
| INITIAL-1 | 2309.66768581353 | 60.083 | 37.38 |
| INITIAL-2 | 2310.6341993138 | 59.885 | 37.38 |
| BLANK-2 | 2312.66284694853 | 59.917 | 37.4 |
| POC-201 | 2302.40007114124 | 59.624 | 37.4 |
| ACR-244 | 2277.31062313817 | 59.786 | 37.39 |
| POR-235 | 2288.18858541504 | 59.525 | 37.41 |
| ACR-229 | 2272.13489556652 | 59.84 | 37.38 |
| POR-245 | 2268.37232119263 | 60.317 | 37.43 |
Data and notes for processing E5 samples TP2, Run 5 20210511
Notes
- Had to completed two seperate titration runs. All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210511 | 2228.879 | 2224.47 | 0.198204516 | 180 | CRM opened 20210418 |
Total Alkalinity Output - Titration Run 1 (8 samples)
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 2023.62860758224 | 60.323 | 35 |
| ACR-360 | 2299.24809778777 | 59.666 | 37.3 |
| POR-355 | 2272.98399534722 | 60.365 | 37.29 |
| POC-394 | 2331.64157338251 | 59.807 | 37.3 |
| ACR-351 | 2308.62561873151 | 59.796 | 37.27 |
| BLANK-5 | 2341.16654731596 | 59.832 | 37.3 |
| INITIAL-1 | 2349.40134517588 | 60.137 | 37.3 |
| ACR-368 | 2335.36944848248 | 60.295 | 37.32 |
| POR-349 | 2312.55677108142 | 59.746 | 37.28 |
Total Alkalinity Output - Titration Run 2 (4 samples)
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 2020.61803398065 | 60.11 | 35 |
| POR-367 | 2271.59356605544 | 59.959 | 37.32 |
| POR-353 | 2246.61383181666 | 60.357 | 37.32 |
| POC-359 | 2320.53306339006 | 59.601 | 37.29 |
| INITIAL-2 | 2354.906625 | 59.539 | 36.52 |
Data and notes for processing E5 samples TP2, remaining Run 1 samples and 1 run 5 sample 20210525
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210525 | 2223.505163 | 2224.47 | -0.043373783 | 180 | CRM opened 20210418 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 2021.113958 | 59.52 | 35 |
| INITIAL-1 | 2328.815162 | 59.78 | 36.1 |
| POR-214 | 2166.262087 | 59.875 | 36.12 |
| POC-200 | 2257.931561 | 60.457 | 36.08 |
| POC-255 | 2290.17382 | 60.446 | 36.1 |
| ACR-237 | 2282.117545 | 59.516 | 36.11 |
| POR-253 | 2121.493182 | 59.948 | 36.09 |
| ACR-258 | 2263.09945 | 59.723 | 36.12 |
| INITIAL-2 | 2354.906625 | 59.539 | 36.52 |
Data and notes for processing E5 samples TP2, remaining Run 2 samples (4), 9 samples from run 3 20210526
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub. Started to record the temperature of seawater in the room for each day. Today it was 24.8°C.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210526 | 2223.47 | 2224.47 | -0.044954528 | 180 | CRM opened 20210418 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| POC-257 | 2294.21281589112 | 60.182 | 36.79 | 2 |
| POR-240 | 2253.77127801894 | 60.261 | 36.88 | 2 |
| POR-262 | 2248.02673011785 | 59.802 | 36.87 | 2 |
| POC-254 | 2287.41202551437 | 60.339 | 36.86 | 2 |
| ACR-265 | 2283.76004008528 | 59.977 | 37 | 3 |
| POC-239 | 2295.81472407995 | 60.457 | 37.03 | 3 |
| INITIAL-1 | 2315.48416509178 | 59.894 | 37.02 | 3 |
| POR-209 | 2254.08211275199 | 60.23 | 37.02 | 3 |
| POC-217 | 2299.82917108649 | 60.113 | 37.01 | 3 |
| BLANK-3 | 2308.18866510965 | 59.689 | 37.02 | 3 |
| POR-221 | 2172.68917435377 | 60.417 | 37.01 | 3 |
| POC-259 | 2285.3899644917 | 59.997 | 37.04 | 3 |
| POC-207 | 2299.9880480591 | 59.653 | 37.04 | 3 |
Data and notes for processing E5 samples TP2, remaining Run 3 samples (3), 5 samples from run 4 20210527
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub. Temperature of room seawater: 24.5°C.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210527 | 2221.347 | 2224.47 | -0.140392992 | 180 | CRM opened 20210418 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| JUNK1 | 2048.4918806143 | 60.049 | 35 | - |
| POR-236 | 2210.78794910016 | 59.592 | 35.35 | 3 |
| POR-206 | 2169.18082109917 | 60.638 | 35.38 | 3 |
| INITIAL-2 | 2314.88140617624 | 60.075 | 35.35 | 3 |
| POR-260 | 2215.42467558492 | 59.651 | 35.5 | 4 |
| POC-222 | 2294.11805543367 | 59.754 | 35.49 | 4 |
| POR-242 | 2002.10749874344 | 60.135 | 35.5 | 4 |
| POR-266 | 2128.69485050727 | 60.084 | 35.49 | 4 |
| POC-248 | 2291.29416890313 | 60.283 | 35.5 | 4 |
Data and notes for processing E5 samples TP2, remaining Run 4 samples (7), 9 samples from run 6 20210601
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub. Temperature of room seawater: 24.2°C.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210601 | 2222.281343 | 2224.47 | -0.098390052 | 180 | CRM opened 20210526 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| INITIAL-2 | 2323.47645891917 | 60.309 | 35.66 | 4 |
| BLANK-4 | 2323.38824894761 | 60.327 | 35.64 | 4 |
| POC-219 | 2314.63393459143 | 60.164 | 35.64 | 4 |
| POR-216 | 2162.56151837602 | 59.636 | 35.64 | 4 |
| INITIAL-1 | 2302.12915601656 | 59.576 | 35.66 | 4 |
| POC-205 | 2306.82987290008 | 60.252 | 35.64 | 4 |
| POR-383 | 2292.09784313125 | 59.837 | 35.83 | 6 |
| POR-381 | 2268.80549000996 | 60.483 | 35.84 | 6 |
| POR-341 | 2303.485165 | 59.517 | 35.82 | 6 |
| POC-238 | 2290.323051 | 59.502 | 35.82 | 6 |
| BLANK-6 | 2330.885054 | 59.868 | 35.81 | 6 |
| POC-378 | 2317.066965 | 60.182 | 35.83 | 6 |
| POC-366 | 2319.944227 | 59.805 | 35.8 | 6 |
| INITIAL-1 | 2335.442874 | 59.799 | 35.84 | 6 |
| ACR-396 | 2313.47919 | 59.592 | 35.85 | 6 |
| INITIAL-2 | 2369.881136 | 59.78 | 35.82 | 6 |
Data and notes for processing E5 samples TP2, remaining Run 6 samples (3), 12 samples from run 7 20210624
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub. Added temperature probe to titrator, continuous temperature being measured during runs.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210624 | 2228.452 | 2224.47 | 0.179008932 | 180 | CRM opened 20210526 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| JUNK1 | 2083.83581489909 | 59.875 | 35 | NA |
| ACR-350 | 2313.45289533546 | 59.604 | 37.56 | 6 |
| ACR-343 | 2312.10286349871 | 60.376 | 37.58 | 6 |
| POC-391 | 2308.18425528784 | 60.361 | 37.7 | 7 |
| BK-7 | 2329.54336354651 | 59.908 | 37.66 | 7 |
| POC-377 | 2318.93041886119 | 59.958 | 37.66 | 7 |
| ACR-347 | 2287.77157182072 | 60.403 | 37.66 | 7 |
| POR-357 | 2301.85458181472 | 59.963 | 37.61 | 7 |
| POR-384 | 2068.74678641006 | 60.493 | 37.61 | 7 |
| POC-371 | 2333.22335273167 | 59.653 | 37.65 | 7 |
| POC-373 | 2319.57500582734 | 59.598 | 37.61 | 7 |
| INITIAL-1 | 2332.42150299334 | 59.664 | 37.68 | 7 |
| POR-338 | 2296.40931234685 | 60.389 | 37.66 | 7 |
| ACR-364 | 2313.9874371687 | 59.822 | 37.65 | 7 |
| INITIAL-2 | 2331.80262384803 | 59.961 | 37.66 | 7 |
Data and notes for processing E5 samples TP2, Run 8 samples (12), 1 samples from run 9 20210706
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210706 | 2230.091863 | 2224.47 | 0.25272822 | 180 | CRM opened 20210526 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| JUNK1 | 2127.87573977338 | 59.736 | 35 | NA |
| POR-365 | 2302.22568628682 | 60.488 | 37.66 | 8 |
| POC-369 | 2332.48754752583 | 59.741 | 37.64 | 8 |
| INITIAL-2 | 2338.97139177075 | 59.892 | 37.61 | 8 |
| POR-340 | 2267.76065570345 | 59.596 | 37.64 | 8 |
| BLANK-8 | 2334.95502705341 | 60.035 | 37.63 | 8 |
| POR-385 | 2268.85278894355 | 59.742 | 37.65 | 8 |
| POC-372 | 2321.65688763208 | 59.923 | 37.68 | 8 |
| POC-375 | 2303.3815103305 | 60.203 | 37.65 | 8 |
| POC-346 | 2325.31511305514 | 59.651 | 37.77 | 8 |
| ACR-393 | 2319.92989417533 | 60.172 | 37.72 | 8 |
| INITIAL-1 | 2336.42991421746 | 60.005 | 37.74 | 8 |
| POC-395 | 2316.5050298392 | 59.603 | 37.74 | 8 |
| POC-48 | 2325.01226734367 | 59.609 | 37.75 | 9 |
Data and notes for processing E5 samples TP2, Run 9 samples (8) 20210708
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210708 | 2228.957083 | 2224.47 | 0.201714718 | 180 | CRM opened 20210526 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| JUNK1 | 2265.60200372629 | 59.841 | 35 | NA |
| POC-42 | 2322.61351513766 | 60.339 | 37.88 | 9 |
| POR-73 | 2216.31310048821 | 60.323 | 37.96 | 9 |
| ACR-186 | 2294.35842028192 | 60.143 | 37.95 | 9 |
| BLANK-9 | 2340.47696082826 | 60.213 | 37.97 | 9 |
| ACR-150 | 2284.40662336857 | 60.464 | 37.95 | 9 |
| INITIAL-1 | 2348.49847005759 | 59.952 | 37.89 | 9 |
| ACR-173 | 2267.79743612355 | 59.932 | 37.95 | 9 |
| POC-50 | 2329.21094044675 | 60.098 | 37.9 | 9 |
Data and notes for processing E5 samples TP2, Run 9 samples (2), run 10 samples (12), and run 11 samples (2) 20210715
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210715 | 2230.460292 | 2224.47 | 0.269290768 | 180 | CRM opened 20210526 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| JUNK1 | 2270.28371447114 | 60.173 | 35 | NA |
| POC-386 | 2324.0085661087 | 60.373 | 38.06 | 9 |
| POR-354 | 2309.65549162679 | 59.868 | 38.06 | 9 |
| INITIAL-2 | 2336.08593365965 | 59.517 | 38.15 | 10 |
| INITIAL-1 | 2334.68461218021 | 60.175 | 38.14 | 10 |
| POC-44 | 2316.96398025246 | 59.912 | 38.17 | 10 |
| BLANK-10 | 2336.38727847286 | 60.094 | 38.18 | 10 |
| POR-79 | 2257.74172138601 | 60.193 | 38.17 | 10 |
| POC-45 | 2319.05071812265 | 60.091 | 38.18 | 10 |
| POR-69 | 2164.33223065006 | 59.976 | 38.14 | 10 |
| POC-68 | 2321.23543991343 | 59.537 | 38.16 | 10 |
| POR-71 | 2184.29178281424 | 60.46 | 38.17 | 10 |
| ACR-139 | 2297.47202192576 | 59.631 | 38.17 | 10 |
| POR-77 | 2231.06018551181 | 59.984 | 38.17 | 10 |
| ACR-145 | 2313.15975234594 | 59.812 | 38.17 | 10 |
| POC-55 | 2314.56794666255 | 60.002 | 38.17 | 11 |
| POR-80 | 2243.56993167095 | 59.773 | 38.17 | 11 |
Data and notes for processing E5 samples TP2, Run 11 samples (8), run 20210811
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20210811 | 2221.057299 | 2224.47 | -0.153416354 | 180 | CRM opened 20210720 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 1934.9754608898 | 60.346 | 35 |
| BLANK-11 | 2324.15793070253 | 59.752 | 39.4 |
| POC-56 | 2306.07052131465 | 59.896 | 39.42 |
| POC-40 | 2296.8754717866 | 60.336 | 39.42 |
| INITIAL-1 | 2336.55412418495 | 59.76 | 39.41 |
| ACR-178 | 2269.5758792525 | 59.807 | 39.42 |
| POR-78 | 2159.98302587912 | 60.382 | 39.41 |
| POR-70 | 2181.20006133922 | 59.548 | 39.42 |
| ACR-190 | 2284.78864861159 | 59.863 | 39.44 |
Data and notes for processing E5 samples TP2, Run 11 samples (1), Run 12 samples (7) 20211110
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20211110 | 2220.494702 | 2224.47 | -0.17870765 | 180 | CRM opened 20210720 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity |
|---|---|---|---|
| JUNK1 | 1844.40951345787 | 60.018 | 35 |
| POR-81 | 2279.69144608703 | 59.848 | 39.33 |
| POR-74 | 2287.49989551604 | 60.48 | 39.42 |
| Initial-1 | 2328.69668652401 | 60.174 | 39.59 |
| POC-41 | 2298.07620701969 | 60.024 | 39.44 |
| POC-47 | 2307.75218797885 | 60.395 | 39.51 |
| BK-12 | 2328.61428528263 | 60.067 | 39.43 |
| POR-76 | 2155.56493361319 | 60.179 | 39.53 |
| ACR-140 | 2277.19768756345 | 59.914 | 39.75 |
Data and notes for processing E5 samples TP2, Run 12 samples (5), Run 13 samples (8) 20220210
Notes
- All samples looked good. Updated timeseries data to urol-e5 GitHub.
Data
CRM Accuracy Data
| Date | CRM value | Batch value | % off | Batch # | Notes |
|---|---|---|---|---|---|
| 20220210 | 2228.61 | 2224.47 | 0.186111748 | 180 | CRM OPENED 20220118 |
Total Alkalinity Output
| SampleID | TA | Mass | Salinity | Run |
|---|---|---|---|---|
| POR_82 | 2274.19295228573 | 60.378 | 40.55 | 12 |
| POC_57 | 2319.26422775354 | 59.96 | 40.76 | 12 |
| INITIAL2 | 2340.15165922546 | 60.448 | 40.73 | 12 |
| POC_43 | 2318.78430225776 | 60.445 | 40.78 | 12 |
| ACR_51 | 2280.9306575261 | 60.203 | 40.76 | 12 |
| INITIAL2.1 | 2330.66583042499 | 60.354 | 40.55 | 13 |
| INITIAL1 | 2335.34615075794 | 59.857 | 40.69 | 13 |
| BK_13 | 2334.90676472687 | 59.544 | 40.72 | 13 |
| POC-53 | 2318.41849540092 | 60.117 | 40.65 | 13 |
| POR-83 | 2239.15805644992 | 59.85 | 40.61 | 13 |
| POR-72 | 2310.663725872 | 59.733 | 40.65 | 13 |
| POR-75 | 2227.34385113541 | 60.328 | 40.63 | 13 |
| POC-52 | 2171.03862117238 | 60.093 | 40.65 | 13 |
