Zymo-Duet-DNA-RNA-Extraction-Protocol
Zymo Duet DNA/RNA Extraction Protocol for Gametogenesis January 2022 samples
Zymo’s Quick-DNA/RNATM Miniprep Plus Kit kit was modified for our lab.
Based on Putnam Lab protocols written by: E. Chille, K. Wong, and E. Strand.
Goal
Process DNA-RNA extractions for the 12-month Gametogenesis time series project in Mo’orea, French Polynesia which investigates the roles of parental effects and epigenetic inheritance in acclimatization and adaptation. The goal of my proposed research is to characterize maternal provisioning, epigenetic inheritance, and cross-generational impacts of thermal stress on Acropora pulchra throughout gametogenesis and early development (i.e., embryo, larval development).
Samples Remaining
January 2022 through August 2022 samples were brought back with CITES permit 20220819 and stored at URI in the flammable fridge (B) and the -80°C freezer (J).
- January 2022, 25 samples remaining (12 processed 20220823)
- February 2022, 36 samples remaining
- March 2022, 40 samples remaining
- April 2022, 40 samples remaining
- May 2022, 40 samples remaining
- June 2022, 40 samples remaining
- July 2022, 40 samples remaining
- August 2022, 40 samples remaining
- September 2022, 40 samples remaining
- October 2022, 40 samples remaining
- November 2022, 40 samples remaining
- December 2022, 40 samples remaining
Process
- Samples were collected and preserved in DNA-RNA shield in Mo’orea, French Polynesia following the Sample Collection Protocol. Initial samples had 1-2 mL of DNA RNA shield added.
Zymo Duet DNA/RNA Extraction Protocol
12 January 2022 samples processed 20220823
| timepoint | tube.number | collection.date | colony.ID | uL taken |
|---|---|---|---|---|
| January 2022 | 418 | 20220115 | ACR-418 | 300 |
| January 2022 | 422 | 20220115 | ACR-422 | 300 |
| January 2022 | 428 | 20220115 | ACR-428 | 300 |
| January 2022 | 431 | 20220115 | ACR-431 | 300 |
| January 2022 | 432 | 20220115 | ACR-432 | 300 |
| January 2022 | 438 | 20220115 | ACR-438 | 300 |
| January 2022 | 457 | 20220115 | ACR-457 | 300 |
| January 2022 | 458 | 20220115 | ACR-458 | 300 |
| January 2022 | 459 | 20220115 | ACR-459 | 300 |
| January 2022 | 460 | 20220115 | ACR-460 | 300 |
| January 2022 | 464 | 20220115 | ACR-464 | 300 |
| January 2022 | 465 | 20220115 | ACR-465 | 300 |

Reagent Preparation
- Add 96 mL 100% ethanol (104 mL 95% ethanol) to the 24 mL DNA/RNA Wash Buffer concentrate before use. DNA/RNA Wash Buffer included with D7003T (Mini Prep Plus Kit) is supplied ready-to-use and does not require the addition of ethanol prior to use. Check kit contents and instructions to confirm prep steps.
- Reconstitute the lyophilized (freeze-dried) DNase I as indicated on the vial prior to use. Mix by inversion. Store frozen aliquots.
- Reconstitute the lyophilized (freeze-dried) 20 mg Proteinase K with 1040 uL Proteinase K Storage Buffer or lyophilized (freeze-dried) 5 mg Proteinase K with 260 uL Proteinase K Storage Buffer. Vortex to dissolve. Store at -20 °C.
Proteinase K Addition
- Place 300 uL of the DNA-RNA shield aliquot into a new 1.5 mL microcentrifuge tube for extractions.
- Add 30 µl of PK digestion buffer to each sample tube (1:10 ratio of PK Digestion Buffer:Sample).
- Add 15 µl Proteinase K to each sample tube (1:2 ratio of Proteinase K:PK Digestion Buffer).
- Vortex for 5-10 seconds and spin down.
- Set timer for 15 minutes and let samples sit. Start labeling other tubes and organizing on tube racks.
- After 15 minutes, spin the samples down for 3 minutes at 16k rcf.
- After samples are spun down, remove the liquid into new 1.5 mL tubes, be careful not to disturb the pellet.
- Proceed to extraction steps.
DNA Extraction
- Set up yellow DNA spin columns and collection tubes, label appropriately
- Warm elution liquids to 70 degrees °C (10mM Tris HCl pH. 8.0 and RNase free water)
- Add equal volume 1:1 (345 µl) DNA/RNA lysis buffer to each sample tube
- Spin samples down for 1 minute at 16k rcf.
- Add 700 µl (total volume) of sample gently to the yellow DNA spin column
- Centrifuge at 16,000 rcf (g) for 3 minutes
- Important Save the flow through from this step: transfer to a new 1.5mL tube labeled for RNA
- Add 400µl DNA/RNA Prep Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 1 minute
- Discard flow through (Zymo kit waste)
- Add 700 µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 1 minute
- Discard flow through (Zymo kit waste)
- Add 400 µl DNA/RNA Wash Buffer gently to the yellow DNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes 30 seconds
- Discard flow through (Zymo kit waste)
- Transfer yellow columns to new 1.5mL microcentrifuge tubes
- Add 50 µl warmed 10 mM Tris HCl to each yellow DNA column by dripping slowly directly on the filer
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 1 minute
- Repeat steps 18-20 for a final elution volume of 100 µl
- Label tubes, store at -20 °C.
RNA Extraction
- Add equal volume (700 µl) 100% EtOH to the 1.5mL tubes labeled for RNA containing the original yellow column flow through
- Vortex and spin down to mix
- Add 700 µl of that liquid to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 1 minute
- Discard flow through (Zymo kit waste)
- Add 700 µl to the green RNA spin columns (the rest from the 1.5mL RNA tubes)
- Centrifuge at 16,000 rcf (g) for 30 seconds
- Get DNase I from freezer
- Discard flow through (Zymo kit waste)
- Add 400 µl DNA/RNA Wash Buffer gently to each green RNA column
- Centrifuge at 16,000 rcf (g) for 1 minute
- Discard flow through (Zymo kit waste)
- Make DNase I treatment master mix:
- 75µl DNA Digestion buffer x # of samples
- 5µl DNase I x # of samples
- Add 80 µl DNase I treatment master mix directly to the filter of the green RNA columns
- Incubate at room temp for 15 minutes
- Add 400 µl DNA/RNA Prep Buffer gently to each column
- Centrifuge at 16,000 rcf (g) for 1 minute
- Discard flow through (Zymo kit waste)
- Add 700 µl DNA/RNA Wash Buffer gently to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 1 minute
- Discard flow through (Zymo kit waste)
- Add 400 µl DNA/RNA Wash Buffer genetly to the green RNA spin columns
- Centrifuge at 16,000 rcf (g) for 2 minutes and 30 seconds
- Discard flow through (Zymo kit waste)
- Transfer green columns to new 1.5mL microcentrifuge tubes
- Add 50µl warmed DNase/RNase free water to each green RNA column by dripping slowly directly on the filer
- Incubate at room temp for 5 minutes
- Centrifuge at 16,000 rcf (g) for 1 minute
- Repeat steps 25-27 for a final elution volume of 100µl
- Label 1.5mL tubes on ice afterwards, and aliquot 5µl into PCR strip tubes to save for Qubit and Tape Station to avoid freeze-thaw of your stock sample
- Store all tubes in the -80 °C
Extraction Content Analysis
These steps analyze the quantity and quality of the DNA/RNA extracted and may be done on a separate day from the extraction.
DNA/RNA Quantity
Follow Broad Range dsDNA and RNA Qubit protocol to analyze sample ++quantity++. Read all samples twice.
DNA/RNA Quantity - 12 January 2022 Samples
| tube.number | colony.ID | qubit.reading.1 | qubit.reading.2 | quibit.average | sample.type | timepoint |
|---|---|---|---|---|---|---|
| DNA_standard_1 | NA | 200.58 | NA | 200.58 | DNA.STD | NA |
| DNA_standard_2 | NA | 24151 | NA | 24151 | DNA.STD | NA |
| 1 | ACR-418 | 25.6 | 25.4 | 25.5 | DNA | JANUARY |
| 2 | ACR-422 | 24 | 23.4 | 23.7 | DNA | JANUARY |
| 4 | ACR-431 | 25.4 | 24.8 | 25.1 | DNA | JANUARY |
| 3 | ACR-428 | 19.8 | 19.3 | 19.55 | DNA | JANUARY |
| 5 | ACR-432 | 15.1 | 14.8 | 14.95 | DNA | JANUARY |
| 6 | ACR-438 | 25 | 24.4 | 24.7 | DNA | JANUARY |
| 7 | ACR-457 | 27.6 | 27 | 27.3 | DNA | JANUARY |
| 8 | ACR-458 | 29.8 | 29 | 29.4 | DNA | JANUARY |
| 9 | ACR-459 | 13.1 | 13 | 13.05 | DNA | JANUARY |
| 10 | ACR-460 | 15.6 | 15.1 | 15.35 | DNA | JANUARY |
| 11 | ACR-464 | 19.2 | 18.8 | 19 | DNA | JANUARY |
| 12 | ACR-465 | 14 | 13.5 | 13.75 | DNA | JANUARY |
| RNA_standard_1 | NA | 422.43 | NA | 422.43 | RNA.STD | NA |
| RNA_standard_2 | NA | 9892.36 | NA | 9892.36 | RNA.STD | NA |
| 1 | ACR-418 | 31.6 | 31.6 | 31.6 | RNA | JANUARY |
| 2 | ACR-422 | 21.6 | 22 | 21.8 | RNA | JANUARY |
| 3 | ACR-428 | 27.6 | 27.6 | 27.6 | RNA | JANUARY |
| 4 | ACR-431 | NA | NA | NA | RNA | JANUARY |
| 5 | ACR-432 | 16.8 | 16.4 | 16.6 | RNA | JANUARY |
| 6 | ACR-438 | 15.2 | 14.4 | 14.8 | RNA | JANUARY |
| 7 | ACR-457 | 18.4 | 18.2 | 18.3 | RNA | JANUARY |
| 8 | ACR-458 | 15.2 | 15.2 | 15.2 | RNA | JANUARY |
| 9 | ACR-459 | 17.4 | 16.6 | 17 | RNA | JANUARY |
| 10 | ACR-460 | 20.4 | 20.6 | 20.5 | RNA | JANUARY |
| 11 | ACR-464 | 13.8 | 13.8 | 13.8 | RNA | JANUARY |
| 12 | ACR-465 | 13.2 | 13 | 13.1 | RNA | JANUARY |
We noticed that the RNA quantity for sample 431 was not detected, this could mean that there was possible contamination during the extraction process, or prior issues when the sample was taken that may have caused this. This sample will need to re-extracted and troubleshooted.
DNA Quality
If DNA quantity is sufficient (typically >10 ng/µL) follow the PPP Agarose Gel Protocol to determine DNA quality. “Good” DNA should form a distinct band a the very top of the gel. See example below:
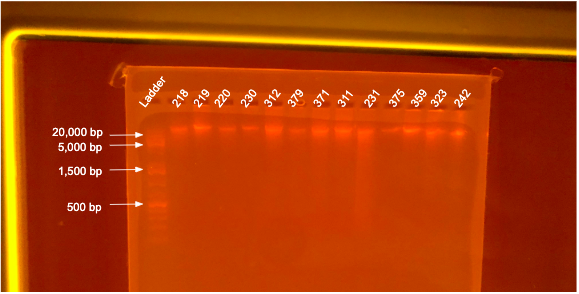
DNA Quality - 12 January 2022 Samples

- Good DNA quality with strong and distinct bands at the top of gel.
RNA Quality
If RNA quantity is sufficient follow the Tape Station Protocol to determine RNA quality and obtain a RNA Integrity Number (RIN). “Good” RNA should have a RIN above 8.0 and form two distinct peaks at the 18S and 28S locations. See example below:
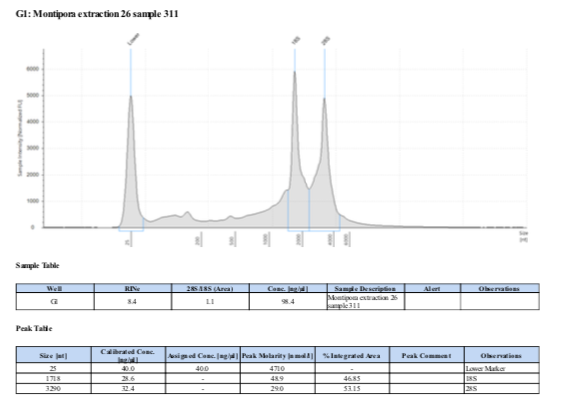
RNA Quality - 12 January 2022 Samples
You can also run RNA quality with a gel, which was done for our samples.
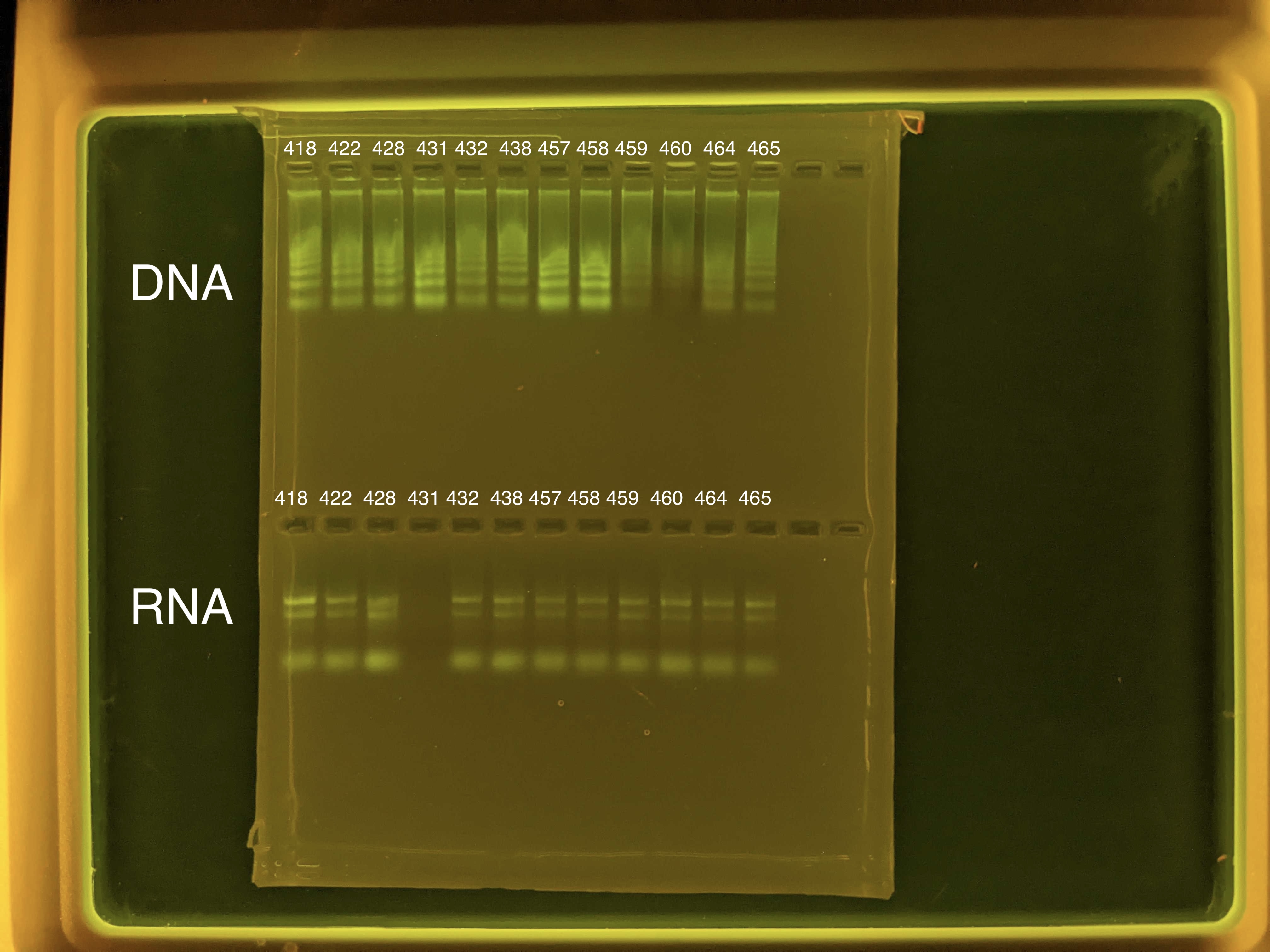
We noticed that the RNA quality for sample 431 was not detected, this could mean that there was possible contamination during the extraction process, or prior issues when the sample was taken that may have caused this. This sample will need to re-extracted and troubleshooted.
