PocHistone Sanger Sequencing Prep Protocol for 22 Remaining Samples
Protocol for PocHistone Sanger Sequencing Prep for the Remaining 22 Samples
Goal:
Amplify the histone 3 region of Pocillopora coral DNA using PCR (polymerase chain reaction). Then use amplification product for Sanger Sequencing and determination of Pocillopora coral species between P. eydouxi and P. meandrina on the remaining 28 samples out of 32 samples. Primer sequences and concept developed in Johnston et. al 2018.
Information on Sanger sequencing at the URI GSC is here. Sequencing takes place after 10am on Tuesdays and Thursdays.
Process
Followed the PocHistone Protocol exactly. See that for in depth details for this protocol.
DNA Dilutions 2020-08-27
DNA dilutions for samples completed by Maggie on 2020-08-27. See the notebook post for in depth details for this protocol.
Histone Amplifications 2021-01-25
- 32 samples plus three negative controls is 35 reactions, use an additional 3 for error
- Made a master mix for 38 samples and 3 reactions each:
- 1900ul Phusion master mix
- 49.4ul working stock PocHistoneF 10uM Primer
- 49.4ul working stock PocHistoneR 10uM Primer
- 1672ul nuclease free water
- Added 97ul of master mix into 35 wells in a new plate
- Used a multichannel to add 3ul of DNA from the dilution plate in to the same orientation wells in the plate with the master mix
- Covered plate and vortexed and spun down
- Separated plate out 2 times into 3 separated reaction mixes each with 33ul
- Covered plates, spun down, and placed in three thermocyclers PocHistone program
1X Bead Cleanup and Quantification 2021-01-25
- Combined triplicate reactions back together
- Added 1X (100ul) beads to each well
- Followed bead cleanup protocol
- Resuspended and eluted DNA in 50ul ultra-pure water and removed into a new plate (same orientation)
Broad Range Qubit 2020-01-25
- dsDNA broad range Qubit assay for 35 samples (n# 40)
- Followed qubit protocol
- Included 32 samples, 3 controls, and 2 standards
| Sample.ID | Qubit reading 1 (ng/ul) | Qubit reading 2 (ng/ul) | average DNA (ng/ul) |
|---|---|---|---|
| std. 1 | 176.98 | NA | NA |
| std. 1 | 20368.25 | NA | NA |
| E10 | 49 | 49 | 49 |
| C22 | 38.2 | 38.4 | 38.3 |
| E10 | 49 | 49 | 49 |
| E2 | 49 | 49.4 | 49.2 |
| C19 | 44.6 | 44.6 | 44.6 |
| C25 | 49.6 | 49.6 | 49.6 |
| C29 | 47.8 | 47.8 | 47.8 |
| C21 | 51.8 | 50.8 | 51.3 |
| E4 | 48.6 | 49.2 | 48.9 |
| E12 | 52.8 | 53.8 | 53.3 |
| E1 | 48 | 47.8 | 47.9 |
| C27 | 52.6 | 53.8 | 53.2 |
| E6 | 49.2 | 49.2 | 49.2 |
| E8 | 44.8 | 45.4 | 45.1 |
| C20 | 54 | 54 | 54 |
| C28 | 52.6 | 52.2 | 52.4 |
| E9 | 54.4 | 54.4 | 54.4 |
| E14 | 52.2 | 51.8 | 52 |
| E15 | 49.6 | 48.4 | 49 |
| C17 | 52.6 | 53 | 52.8 |
| E3 | 47.6 | 47.6 | 47.6 |
| C30 | 55.2 | 55.2 | 55.2 |
| C24 | 45.8 | 45.6 | 45.7 |
| E16 | 48 | 47.6 | 47.8 |
| C31 | 38.4 | 38 | 38.2 |
| E7 | 44.8 | 45.4 | 45.1 |
| C23 | 43.6 | 43.6 | 43.6 |
| E11 | 45.8 | 45.8 | 45.8 |
| E13 | 40 | 40 | 40 |
| C18 | 35 | 35.2 | 35.1 |
| E5 | 43.6 | 43.8 | 43.7 |
| C32 | 33 | 33.4 | 33.2 |
| C26 | 32.8 | 32.8 | 32.8 |
1% Gel To Confirm Bands 2021-01-26
- Made a 1% gel in the medium gel box
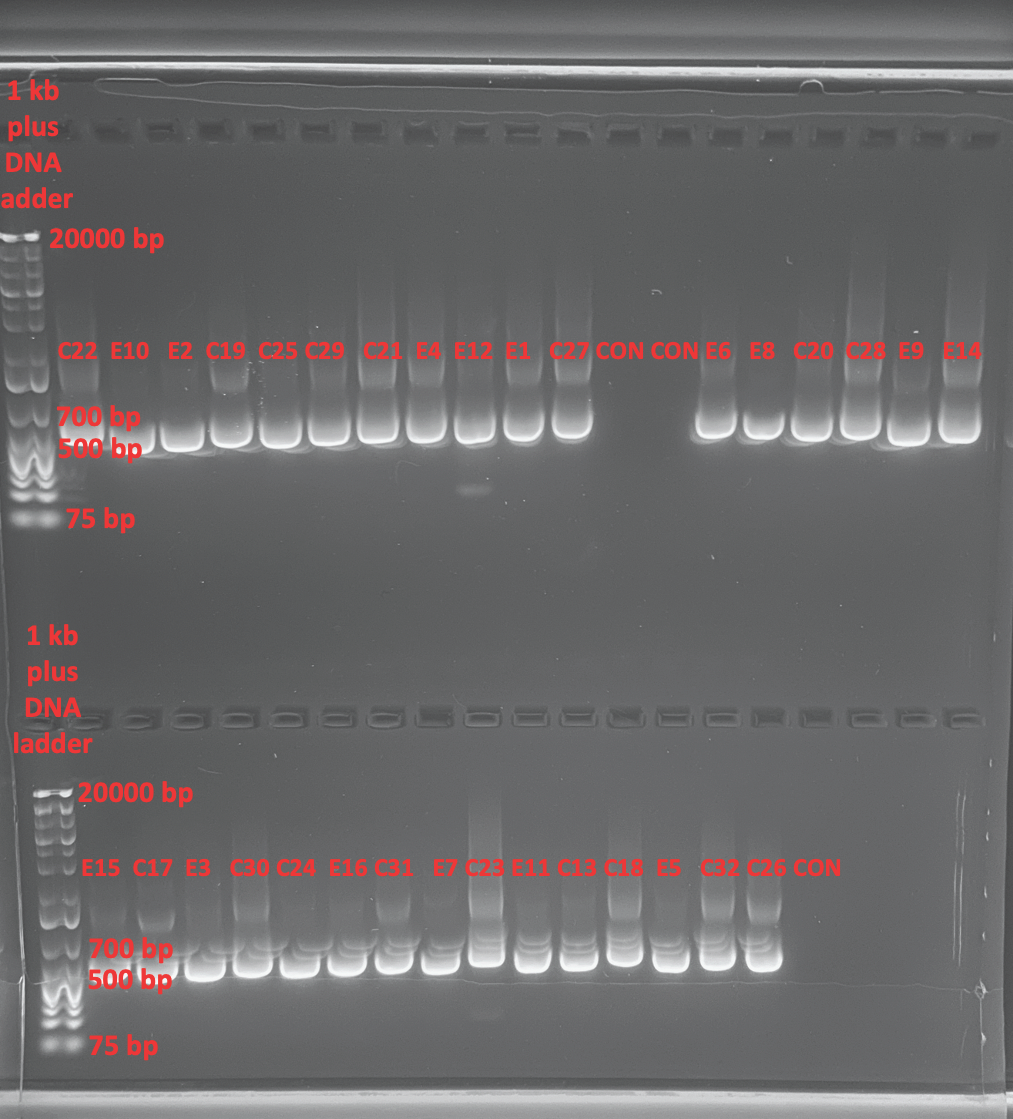
- Bands are strong but some smearing is noticeable which could be an issue. It’s not contamination because our controls are blank. So it’s either an imaging artifact or something that amplified non-specific in the sample. We will be sequencing four samples from a sample size of each treatment and one with smearing and one without to see if there is more of a problem that needs troubleshooting.
Dilution (2021-01-26) and Sequencing Prep for 22 Samples Redone with Reverse Primer (2021-03-08)
All samples were diluted at the same time on 2021-01-26
- Prepped the 22 samples that needed to be redone for sequencing
- For sequencing I needed 45ul (22 * 2 +1 for error) of 3.2uM PocHistone R primer
- The PocHistone product should be ~669 bp, so 669 / 100 * 1.25 * 2 = 16.7ng of each sample
- 16.7ng is still less than 1ul for every sample, so a 1:10 dilution was done for each sample on 2021-01-26
- Added 18ul of ultra pure water to the 96-well plate for 32 samples, and added 2ul of each cleaned DNA to their corresponding well
- Pipetted 16.7ng of each sample from diluted stock to the 96-well plate for 32 samples
- Increased volume in tubes to 10ul with ultra pure water
| sample ID | GSC sample code | dilution.conc | ng.needed | volume diluted PCR product | volume ultra pure H2O |
|---|---|---|---|---|---|
| C22 | HPD65 | 3.83 | 16.7 | 4.36 | 5.64 |
| E2 | HPD66 | 4.92 | 16.7 | 3.39 | 6.61 |
| C19 | HPD67 | 4.46 | 16.7 | 3.74 | 6.26 |
| C29 | HPD68 | 4.78 | 16.7 | 3.49 | 6.51 |
| C21 | HPD69 | 5.13 | 16.7 | 3.26 | 6.74 |
| E4 | HPD70 | 4.89 | 16.7 | 3.42 | 6.58 |
| E1 | HPD71 | 4.79 | 16.7 | 3.49 | 6.51 |
| C27 | HPD72 | 5.32 | 16.7 | 3.14 | 6.86 |
| C20 | HPD73 | 5.4 | 16.7 | 3.09 | 6.91 |
| C28 | HPD74 | 5.24 | 16.7 | 3.19 | 6.81 |
| E9 | HPD75 | 5.44 | 16.7 | 3.07 | 6.93 |
| E14 | HPD76 | 5.2 | 16.7 | 3.21 | 6.79 |
| E15 | HPD77 | 4.9 | 16.7 | 3.41 | 6.59 |
| E3 | HPD78 | 4.76 | 16.7 | 3.51 | 6.49 |
| E16 | HPD79 | 4.78 | 16.7 | 3.49 | 6.51 |
| C31 | HPD80 | 3.82 | 16.7 | 4.37 | 5.63 |
| C23 | HPD81 | 4.36 | 16.7 | 3.83 | 6.17 |
| E11 | HPD82 | 4.58 | 16.7 | 3.65 | 6.35 |
| C18 | HPD83 | 3.51 | 16.7 | 4.76 | 5.24 |
| E5 | HPD84 | 4.37 | 16.7 | 3.82 | 6.18 |
| C32 | HPD85 | 3.32 | 16.7 | 5.03 | 4.97 |
| C26 | HPD86 | 3.28 | 16.7 | 5.09 | 4.91 |
Sequencing prep on 2021-03-01
- Added 16.7 ng of DNA following the volume of the diluted PCR product for each sample above to each corresponding well in a 96-well plate
- Added the volume of ultra pure H2O corresponding to each sample above into the 96-well plate wells
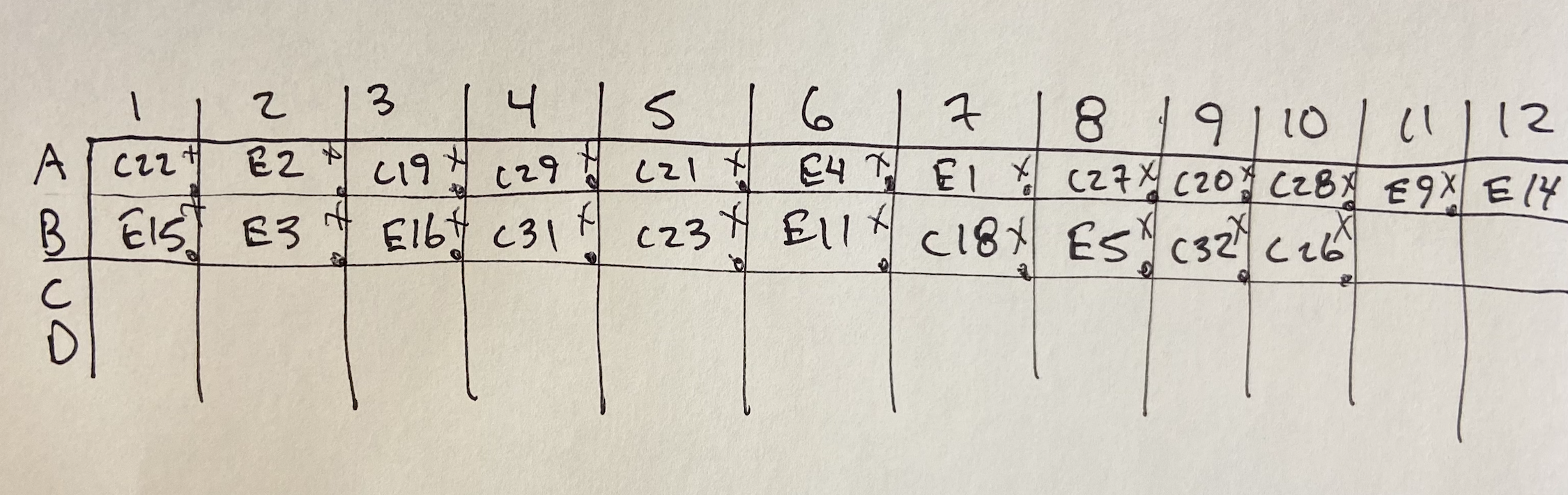
- Made 45ul of diluted 10uM stock PocHistone R to 3.2uM
- Added 30.6ul of ultra pure water to 1 PCR tube
- Added 14.40ul of 10uM PocHistone R primer to tube
- Vortexed and centrifuged to mix
- Added 2ul of 3.2uM primer to each of the 22 wells of the 96-well plate
Sequencing Submission 2021-03-08
- Submitted remaining 22 samples for processing in the GSC lab at 2:00 pm
Spreadsheet for Sequencing
| Sample IDa | Well (GSC use only) | Template Typeb | A. Template Size (bases) | B. Template Stock Conc. (ng/µl) | C. PCR template: ng needed = ((A ÷ 100) x 1.25) x 2 | D. PCR template: Volume = (C ÷ B) µl | F. Volume PCR-H20 needed (10 minus D or E) µl | G. Volume primer needed 1 µl per reaction |
|---|---|---|---|---|---|---|---|---|
| HPD65 | PCR | 669 | 3.83 | 16.725 | 4.36 | 5.64 | 2 | |
| HPD66 | PCR | 669 | 4.92 | 16.725 | 3.39 | 6.61 | 2 | |
| HPD67 | PCR | 669 | 4.46 | 16.725 | 3.74 | 6.26 | 2 | |
| HPD68 | PCR | 669 | 4.78 | 16.725 | 3.49 | 6.51 | 2 | |
| HPD69 | PCR | 669 | 5.13 | 16.725 | 3.26 | 6.74 | 2 | |
| HPD70 | PCR | 669 | 4.89 | 16.725 | 3.42 | 6.58 | 2 | |
| HPD71 | PCR | 669 | 4.79 | 16.725 | 3.49 | 6.51 | 2 | |
| HPD72 | PCR | 669 | 5.32 | 16.725 | 3.14 | 6.86 | 2 | |
| HPD73 | PCR | 669 | 5.4 | 16.725 | 3.09 | 6.91 | 2 | |
| HPD74 | PCR | 669 | 5.24 | 16.725 | 3.19 | 6.81 | 2 | |
| HPD75 | PCR | 669 | 5.44 | 16.725 | 3.07 | 6.93 | 2 | |
| HPD76 | PCR | 669 | 5.2 | 16.725 | 3.21 | 6.79 | 2 | |
| HPD77 | PCR | 669 | 4.9 | 16.725 | 3.41 | 6.59 | 2 | |
| HPD78 | PCR | 669 | 4.76 | 16.725 | 3.51 | 6.49 | 2 | |
| HPD79 | PCR | 669 | 4.78 | 16.725 | 3.49 | 6.51 | 2 | |
| HPD80 | PCR | 669 | 3.82 | 16.725 | 4.37 | 5.63 | 2 | |
| HPD81 | PCR | 669 | 4.36 | 16.725 | 3.83 | 6.17 | 2 | |
| HPD82 | PCR | 669 | 4.58 | 16.725 | 3.65 | 6.35 | 2 | |
| HPD83 | PCR | 669 | 3.51 | 16.725 | 4.76 | 5.24 | 2 | |
| HPD84 | PCR | 669 | 4.37 | 16.725 | 3.82 | 6.18 | 2 | |
| HPD85 | PCR | 669 | 3.32 | 16.725 | 5.03 | 4.97 | 2 | |
| HPD86 | PCR | 669 | 3.28 | 16.725 | 5.09 | 4.91 | 2 |
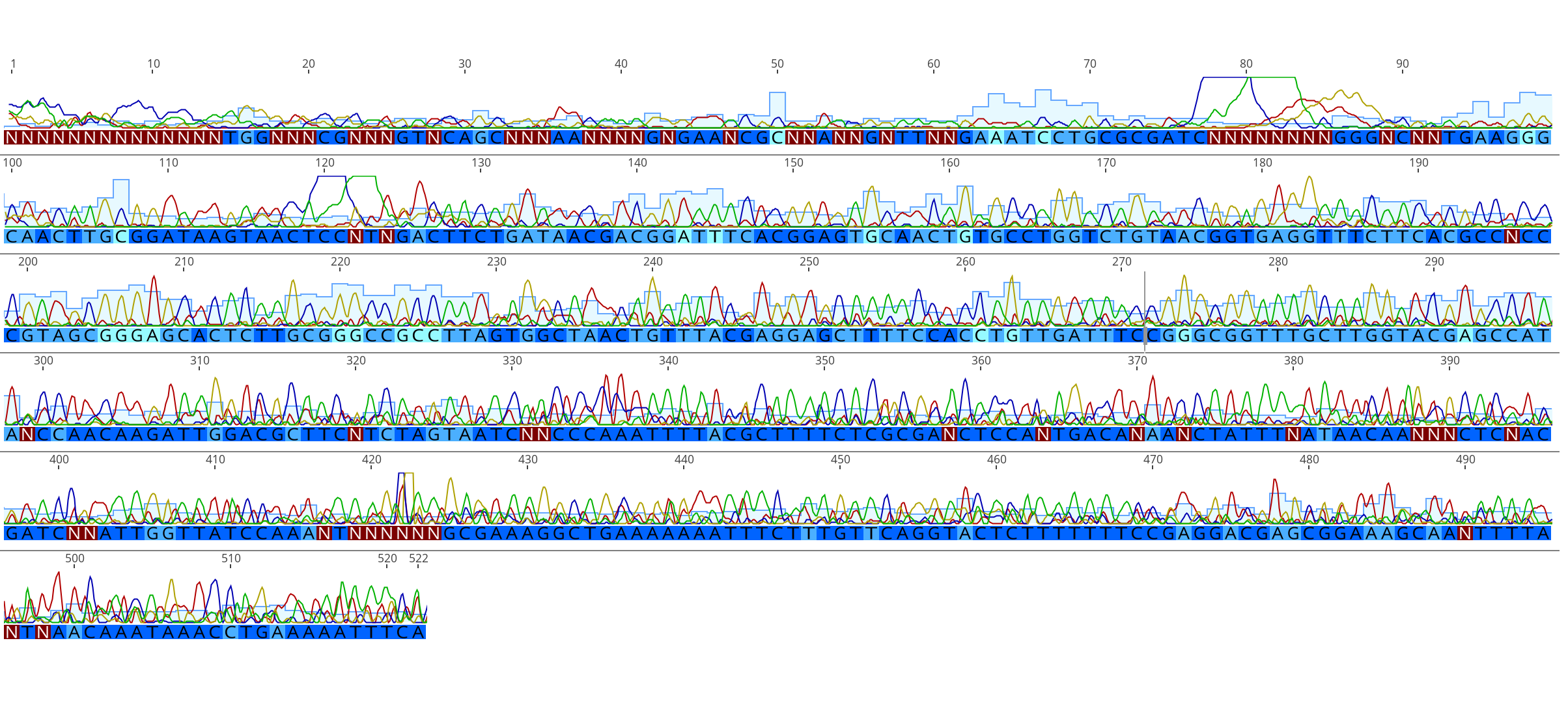
Sequences Received 2021-03-09
- All sequences good besides sample HPD81, sample C23